The SARS-COV-2 virus responsible for the ongoing Covid-19 pandemic, which is progressively spreading throughout the world, belongs to the coronavirus family, the first members of which were identified almost a century ago [1]. These enveloped viruses possess an RNA (ribonucleic acid) genome and characteristic surface projections, recognizable by electron microscopy that give them a crown-like aspect (hence their name ‘corona’; Fig. 1). These projections correspond to the surface spike glycoprotein, also known as the S protein, which plays a key role in the entry of the virus into host cells.
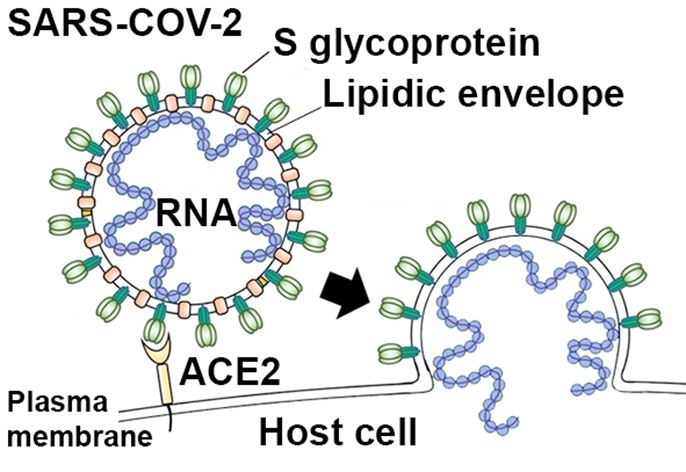
Figure 1: Schematic diagram of SARS-COV-2 virus. Virus entry into a host cell requires the interaction of the S glycoprotein (green) found at the surface of viral particles with a receptor (ACE2; yellow) located at the plasma membrane of the infected cell. This interaction leads to the fusion of the viral envelope with the plasma membrane of the host cell, thus allowing the virus to enter the cell and release its RNA genome (blue) that will serve for the production of new viral particles.
For several weeks already, the medical and scientific community worldwide has been strongly committed to improving our understanding of the specificities of this new virus, SARS-COV-2, and to finding new therapeutic and protective strategies to put an end to this pandemic. Among these researchers, structural biologists have been playing an essential role in determining the atomic-scale structures of the main SARS-COV-2 proteins by X-ray crystallography and/or cryo-electron microscopy (cryo-EM). Their structural studies focus largely on two key viral proteins: the S glycoprotein and a protease (an enzyme capable of cutting other proteins) essential to the virus, responsible for the production of several viral proteins from a single polyprotein.
On the surface of the virus, the S glycoprotein allows the virus to enter human cells via its interaction with a receptor, identified as the enzyme ACE2, present on the surface of infected cells [2]. During the month of March 2020, several partial structures of the SARS-COV-2 S protein, alone or in complex with its receptor ACE2, were determined by cryo-EM, thereby revealing its mode of receptor recognition and the molecular mechanisms underlying the fusion of the viral envelope with the plasma membrane of infected cells [2,3,4]. This structural information can now be exploited firstly to identify inhibitors of these key stages of the viral cycle, which could ultimately serve as antiviral drugs, and, secondly, to develop vaccines. Being on the surface of these viruses, the S glycoprotein plays an essential role in the host immune response. Most neutralizing antibodies recognize this protein. The development of an effective vaccine against SARS-COV-2 will thus require identifying the regions of the S protein displaying the strongest immunogenic potential and capable of inducing an effective and specific immune response against the virus.
A second viral protein, subject of numerous structural studies, is the main protease of SARS-COV-2, called MPRO or 3CLPRO, which currently represents one of the most promising therapeutic targets for the development of antiviral drugs against SARS-COV-2. The first step was the determination of the crystal structure of this enzyme at high resolution, which was achieved in March 2020 [5,6]. This structural data is now being used by numerous research teams to identify specific and effective inhibitors of this protease by theoretical (modelling, in silico drug design etc) [6] and/or experimental approaches, in particular by determining the crystal structures of complexes between the protease and small inhibitory molecules. In this context, large amounts of crystallographic data have been collected by the XChem team from the Diamond Light Source in the UK, following a massive high-throughput screen of small molecules, and a call for volunteers has recently been launched to find crystallographers, structural biologists and chemists who would like to take part in the analysis of these data, in the design of new molecules or in the synthesis of inhibitors.
In France, following the French government measures, the major large instruments, including the European Synchrotron Radiation Facility (ESRF), have been closed since March 17th 2020, but are nonetheless engaged in the fight against Covid-19. The ESRF, for example, is ready to provide exceptional access to its facilities for Covid-19 related projects.
For all researchers wishing to use their expertise to combat Covid-19, you can visit the site: https://crowdfightcovid19.org/. CRCA-CBI researchers in Toulouse have set up this platform to put the skills and expertise of the wider scientific community not presently engaged in Covid-19 research at the service of those already heavily involved in this fight.
Joanna Timmins, vice-president of the French Crystallographic Association – (Institut de Biologie Structurale, Virus Infection & Cancer Group, DNA Damage & Repair Team, Grenoble)
Text adapted from the original French version published on April 3rd, 2020, on the website of the French Crystallography Association.
References
- Peiris J. Coronaviruses. Clinical Virology, Fourth Edition. 2017. p. 1243-1265.
- Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020 Mar 6. pii: S0092-8674(20)30262-2.
- Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020 Mar 27. 367 (6485) : 1444-1448.
- Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, McLellan JS. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020 Mar 13. 367 (6483): 1260-1263.
- Zhang L, Lin D, Sun X, Curth U, Drosten C, Sauerhering L, Becker S, Rox K, Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020 Mar 20. pii : eabb3405.
- Coronavirus SARS-CoV2: BESSY II data accelerate drug development. 2020 March 19.
- Chen YW, Yiu CB, Wong KY. Prediction of the SARS-CoV-2 (2019-nCoV) 3C-like protease (3CL (pro)) structure: virtual screening reveals velpatasvir, ledipasvir, and other drug repurposing candidates. F1000Res. 2020 Feb 21. 9:129.
